

- CONVERTING FROM BIOEDIT TO MEGA FULL
- CONVERTING FROM BIOEDIT TO MEGA REGISTRATION
- CONVERTING FROM BIOEDIT TO MEGA SOFTWARE
- CONVERTING FROM BIOEDIT TO MEGA FREE
- CONVERTING FROM BIOEDIT TO MEGA WINDOWS
These sequences will be aligned with the input sequences above, being used as a constraint. Paste protein or DNA sequences in fasta format.
CONVERTING FROM BIOEDIT TO MEGA FREE
Please let me know if this worked for you and feel free to send me over your fasta file to check it out. Multiple sequence alignment and NJ / UPGMA phylogeny. Just add the gaps manually in notepad, save and your input file is ready for use in network.
CONVERTING FROM BIOEDIT TO MEGA FULL
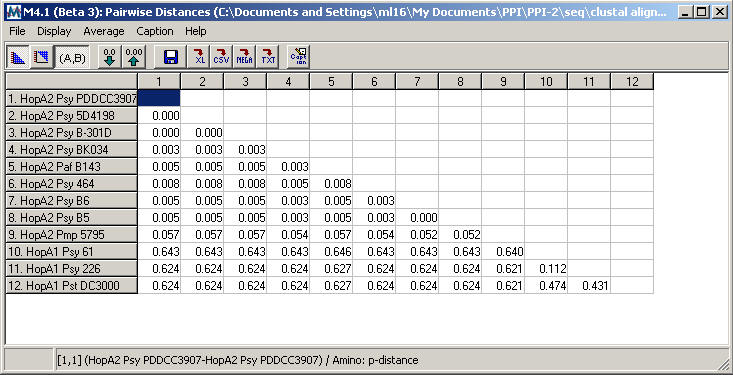
Quick solution: If you do not want to repeat the full path, simply open your nex file in notepad and you will likely (in the end of your alignment) find that some of your sequences are shorter than others usually by one or few gaps. The results is that once you have created your nexus file via the path I suggest, some sequences will be shorter that the length of the alignment and therefore they will not have the specific number of nucleotides reported in the nexus block. ©Northwest Association for Biomedical ResearchUpdated Aug2 Quality values: A quality value is a number that is used to assess the accuracy of each base in a DNA sequence. If the file is only saved as fasta and not exported as alignment, Bioedit will not match the differences of the end missing data in length by adding the final gaps. In the latter case, your sequences, although aligned manually, are of different length because of missing data in their end. You have initially imported your sequences in Bioedit to create your alignment but you haven't exported your fasta file as alignment you have simply saved it as a fasta file. What is likely happening in this case is the following: Hi Pamoda and many thanks for your email. ExampleĪlignment files are from Poore and Andreakis Sure there are other personalized ways to do it. To be always successful and literally based on few clicks of your mouse but I am Or amino acid sequences aligned with… your preferred algorithm. This post I suggest a quick and free mode to create nexus input files for Network starting from DNA

Input files (.rdf) from next generation sequencing files respectively. Despite Network is freely distributed, the add-ons are The use of “recommended” add-ons such as DNA Alignment, Network Publisher and pibase for importingĭata without errors, create publication quality graphics and generating network However, contrary to TCS and Splitstree, the Network team suggests In the past and I was impresed for its versatility, the graphics (compared toĪnd its friendly interface.

The median joining algorithm to generate genealogical trees from DNA,Īminoacid, linguistic and other data but can also provide age estimates for theĪncestral nodes in the tree topology.
CONVERTING FROM BIOEDIT TO MEGA SOFTWARE
provides, free of charge, the phylogenetic Network software in its as of today version Help to people intersted in genealogical network reconstruction under the If you need to assemble multiple overlapping sequence reads, please see ChromasPro.It is a bit of time I was thinking that this post may be of An introductory discount is available by following the link under the Options menu in Chromas.
CONVERTING FROM BIOEDIT TO MEGA REGISTRATION
The PeakTrace RP component requires registration for a free account, and is a paid service after the 40 free units have been used.
Version history.Ĭhromas is free of charge.
CONVERTING FROM BIOEDIT TO MEGA WINDOWS
ab1 files using the PeakTrace RP component from Nucleics Pty Ltd to improve peak readability and extract more high-quality bases.Ĭompatible with Windows XP, Vista, 7, 8, 10.


 0 kommentar(er)
0 kommentar(er)
